Project leader : C. Moali and S. Vadon-Le Goff
Collagen fibril formation is a complex process involving both intracellular and extracellular steps.
- At the intracellular level, polypeptide chains are synthesized and modified with several types of post-translational modifications (hydroxylations, glycosylations, disulfide bridges etc) before assembly into procollagen trimers. The latter step is critical and implies specific chain recognition, with appropriate composition and stoichiometry, to avoid the production of dysfunctional chimeric molecules by cells that synthesize several collagen types. In the case of fibrillar collagen, this recognition mechanism is controlled by the C-terminal domain called C-propeptide.
One of our research interest is to understand the molecular details of collagen C-propeptides governing recognition mechanisms and the structural impact of mutations in these domains known to cause genetic diseases (Osteogenesis Imperfecta, Chondrodysplasias, Ehlers-Danlos etc).
- At the extracellular level, after secretion of procollagen molecules, proteolytic maturation is required to decrease collagen solubility and trigger fibril formation. The main proteinases involved are the BMP-1/tolloid-like proteinases (BTPs), meprins and ADAMTS-2, 3 and 14. BTPs are assisted in this function by a 50 kDa glycoprotein called Procollagen C-Proteinase Enhancer-1 (PCPE-1) which can significantly increase their cleavage efficiencies. Interestingly, PCPE-1 is specific for fibrillar procollagens, as it does not affect other BTP activities, and potentially allows to speed up collagen fibril formation in a significant manner when active collagen deposition is needed (development, tissue repair).
We are trying to understand the mechanism of action of the large proteolytic complexes involved in procollagen maturation with two (protease-substrate, substrate-enhancer, protease-enhancer) or three (protease-substrate-enhancer) partners.
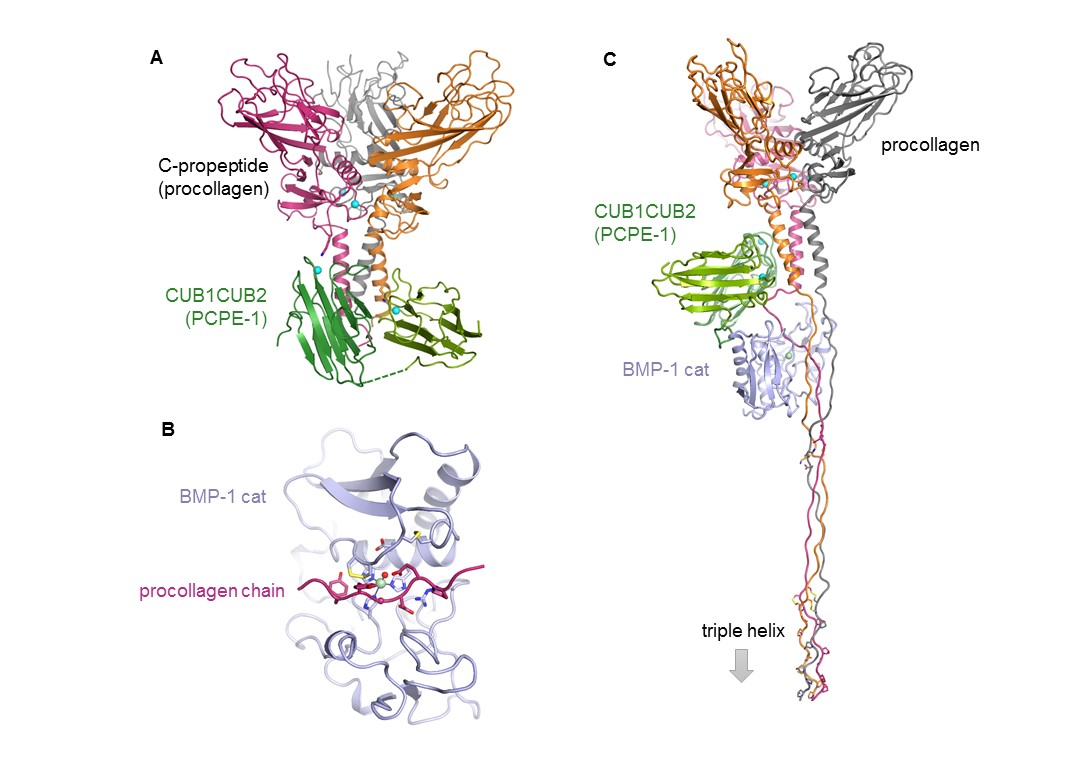
(A) Crystal structure of the complex between the C-propeptide domain of collagen III and the active part of PCPE-1 (CUB1CUB2). (B) Model of one procollagen III chain in BMP-1 catalytic site. (C) Model of the three-partner complex (procollagen III : CUB1CUB2 : BMP-1 cat). (Adapted from Pulido et al. Structure 2018).
Main techniques used
• Protein production and purification
• Enzyme assays
• Surface plasmon resonance / fluorescence / circular dichroism
• Mass spectrometry
• X-ray scattering
• Cryo-electron microscopy
• Crystallography
Selection of publications :
Structural basis of fibrillar collagen trimerization and related genetic disorders. Bourhis JM, Mariano N, Zhao Y, Harlos K, Exposito JY, Jones EY, Moali C, Aghajari N, Hulmes DJ. Nat Struct Mol Biol. 2012, 19(10):1031-6
Procollagen C-proteinase enhancer grasps the stalk of the C-propeptide trimer to boost collagen precursor maturation. Bourhis JM, Vadon-Le Goff S, Afrache H, Mariano N, Kronenberg D, Thielens N, Moali C, Hulmes DJ. Proc Natl Acad Sci U S A. 2013, 110(16):6394-9
Clinical, structural, biochemical and X-ray crystallographic correlates of pathogenicity for variants in the C-propeptide region of the COL3A1 gene. Stembridge NS, Vandersteen AM, Ghali N, Sawle P, Nesbitt M, Pollitt RC, Ferguson DJ, Holden S, Elmslie F, Henderson A, Hulmes DJ, Pope FM. Am J Med Genet A. 2015, 167A(8):1763-72.
Structural basis of homo- and heterotrimerization of collagen I. Sharma U, Carrique L, Vadon-Le Goff S, Mariano N, Georges RN, Delolme F, Koivunen P, Myllyharju J, Moali C, Aghajari N, Hulmes DJ. Nat Commun. 2017, 8:14671.
Structural basis for the acceleration of procollagen processing by procollagen C-proteinase enhancer-1. Pulido D, Sharma U, Vadon-Le Goff S, Hussain SA, Cordes S, Mariano N, Bettler E, Moali C, Aghajari N, Hohenester E, Hulmes DJ. Structure 2018, 26: 1384-92.




